
Nicotinamide N-methyltransferase epigenetic and metabolic rewiring promotes metastatic progression
High-grade serous carcinoma (HGSC) is amongst the most deadly forms of cancer. Approximately 80% of patients with HGSC suffer recurrence following surgical resection and cytoreduction; a consequence of the widespread and inaccessible metastatic lesions throughout the abdominal cavity. HGSC progression and metastases co-evolves with the tumor microenvironment (TME), composing fibroblasts, mesenchymal stem cells, osteoblasts, and chondrocytes. Within the TME, fibroblasts and other normal cells can transition to a tumor promoting phenotype, termed cancer associated fibroblasts (CAFs). CAFs are an abundant cell type that support extracellular matrix remodeling, angiogenesis, and metastasis (1). As a result, CAFs represent a potential therapeutic target to reduce tumor growth and prevent metastasis. However, the factor(s) that regulate the switch to the CAF phenotype remain unknown. In this article, Eckert et al. couple laser-captured microdissection with ultra-high-sensitivity mass-spectrometry-based proteomics to clearly delineate the proteome of HGSC cancer cells and corresponding stroma at the primary and metastatic sites (2). This study provides novel insights into the transcriptomic and epigenetic alterations of metastatic stromal cells and identifies potential therapeutic strategies for combating HGSC.
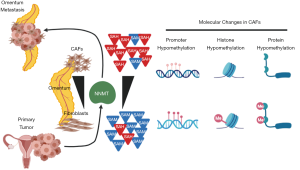
The authors discovered a consistent proteomic signature in the metastatic stroma across all HGSC patients. This was in stark contrast to the proteome of the tumor compartment, which was primarily characterized by patient-specific protein signatures. The most prominent signature was observed in the omentum metastatic samples, where a group of 62 proteins was universally up- or downregulated. Within this gene signature, the authors focused on the upregulated protein nicotinamide N-methyltransferase (NNMT). NNMT transfers the methyl group from S-adenosylmethionine (SAM) to nicotinamide to generate S-adenosyl homocysteine and the inert product 1-methylniotinamide (1-MNA). As a result, NNMT regulates the cellular methylation potential (SAM/SAH) and influences nicotinamide metabolism. NNMT is a key mediator of cell fate through the global shift in methionine metabolism, histone methylation, and protein methylation (3-5). NNMT regulates the naïve-to-primed transition in human embryonic stem cells and promotes the cancer stem cell phenotype (3,6,7). These discoveries are in line with a growing body of evidence that supports the notion that metabolic plasticity may drive cellular reprogramming (8). Therefore, the authors’ hypothesized that NNMT may play a similar role in transitioning fibroblasts to the CAF phenotype.
CAFs differ from normal fibroblasts by altered morphology and increased cytokine secretion. NNMT silencing in CAFs led to a reversion in morphology and decreased CAF marker expression. Further, gene expression analysis demonstrated that NNMT silencing changes the expression of thousands of genes. These differential regulated genes were enriched in CAF markers including a significant number of the differentially regulated proteins identified in the initial proteomic screen. NNMT is known to influence gene expression by altering methionine metabolism and causing histone hypo-methylation (3). The authors investigated whether NNMT induced a similar change in epigenetics through the alteration to the methylation potential. Indeed, NNMT expression was inversely correlated with methylation potential and significantly decreased intracellular nicotinamide concentrations. To determine whether this decreased methylation potential altered histone methylation, the authors quantified the relative concentrations of histone methylation at arginine and lysine residues. NNMT knockdown cells showed increased histone methylation at transcriptional regulation sites H3K4 and H3K27. Conversely, NNMT overexpressing cells showed global decreases in H3K27 occupancy. Importantly, occupancy was found at a significant number of the promoters of NNMT-regulated genes. Enhancer of zeste homolog 2 (EZH2) catalyzes the addition of methyl marks to H3K27. To demonstrate that the decrease in H3K27 occupancy drove the CAF phenotype, EZH2 was inhibited genetically and pharmacologically in NNMT knockdown cells. In all cases, inhibition of EZH2 was sufficient to promote the morphology and markers associated with CAFs. The authors concluded that NNMT alters the transcriptome by altering repressive chromatin marks.
To validate these findings, the authors investigated the role of NNMT expression in fibroblasts in promoting ovarian cancer cell metastasis in vivo. Mouse ovarian cancer cells pre-treated with conditioned media from NNMT overexpressing fibroblasts increased metastasis. Co-injection of HGSC cell with NNMT knockdown fibroblasts reduced proliferation, tumor burden, and stromal H3K27 trimethylation in vivo. In patient tissue, stromal NNMT was prognostic for recurrence-free survival, overall survival, and platinum resistance. In contrast, NNMT expression in the tumor compartment was not prognostic. The authors concluded that NNMT plays a significant role in the tumor- and metastatic-promoting functions of CAFs.
Despite marked genetic and proteomic heterogeneity of epithelial ovarian cancer, the metastatic stromal proteome was notably uniform and characterized by high NNMT expression and consequent epigenetic and transcriptomic rewiring. NNMT metabolically regulates CAF differentiation in the stromal compartment. Stromal NNMT expression depletes universal methyl donor SAM, remodels methylome of histones, resulting with widespread gene expression changes in the tumor stroma. In CAFs, stromal NNMT expression was necessary and sufficient to recapitulate the progressive and metastatic cancer phenotype (Figure 1). Inhibition of NNMT reversed the CAF phenotype suggesting that NNMT inhibition may represent a feasible therapeutic strategy to combat HGSC.
Acknowledgments
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Valkenburg KC, de Groot AE, Pienta KJ. Targeting the tumour stroma to improve cancer therapy. Nat Rev Clin Oncol 2018;15:366-81. [Crossref] [PubMed]
- Eckert MA, Coscia F, Chryplewicz A, et al. Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts. Nature 2019;569:723-8. [Crossref] [PubMed]
- Sperber H, Mathieu J, Wang Y, et al. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat Cell Biol 2015;17:1523-35. [Crossref] [PubMed]
- Ulanovskaya OA, Zuhl AM, Cravatt BF. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat Chem Biol 2013;9:300-6. [Crossref] [PubMed]
- Palanichamy K, Kanji S, Gordon N, et al. NNMT Silencing Activates Tumor Suppressor PP2A, Inactivates Oncogenic STKs, and Inhibits Tumor Forming Ability. Clin Cancer Res 2017;23:2325-34. [Crossref] [PubMed]
- Jung J, Kim LJ, Wang X, et al. Nicotinamide metabolism regulates glioblastoma stem cell maintenance. JCI Insight 2017;2. [Crossref] [PubMed]
- Pozzi V, Sartini D, Rocchetti R, et al. Identification and characterization of cancer stem cells from head and neck squamous cell carcinoma cell lines. Cell Physiol Biochem 2015;36:784-98. [Crossref] [PubMed]
- Folmes CD, Dzeja PP, Nelson TJ, et al. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell 2012;11:596-606. [Crossref] [PubMed]
Cite this article as: Jacob JR, Chakravarti A, Palanichamy K. Nicotinamide N-methyltransferase epigenetic and metabolic rewiring promotes metastatic progression. Biotarget 2019;3:21.


