
A gene expression map of colon tissue in ulcerative colitis: new methods rewrite old stories
Ulcerative colitis (UC) constitutes a chronic relapsing inflammatory disease of the colonic mucosa with increasing incidence in all ages that imparts significant morbidity; As therapeutic options remain limited, UC urges as an important health concern worldwide (1,2). Along with Crohn’s disease (CD) represent the two main types of inflammatory bowel disease (IBD). Besides their phenotypic similarities there are substantial differences between UC and CD. Inflammation pattern in UC is uninterrupted and is restricted to colonic mucosa, while in CD is patchy and may affect any part of the gastrointestinal tract in a transmural fashion often leading to fibrotic complications. These differences probably reflect the distinctive cellular mechanisms and pathways underlining their respective pathogenesis (3,4).
Despite extensive investigations over the last years the pathogenesis of UC remains incompletely understood. It is suggested that complex interactions among genetic, immune, microbial and environmental factors disrupt the homeostatic mechanisms in colonic mucosa leading to disease development. Today, there is a pressing need for a better understanding of UC pathogenesis that will provide novel molecular mechanisms and targets permitting tailored therapeutic approaches to individual patients with UC (1,3,4). To this end, several different approaches have been used in order to nominate candidate molecular pathways in the pathogenesis of inflammation in UC and reveal biomarkers that can predict disease relapse or response to treatment.
In hypothesis-driven studies, several candidate genes, proteins, and/or cell populations have been analyzed at intestinal tissue level or in peripheral blood in an attempt to correlate UC clinical traits with certain molecular characteristics. Over the last years, these approaches have proposed a variety of molecular mechanisms offering potential models responsible for gut inflammation in UC (3,4); however they did not fully elucidate the pathophysiology of this complex disease. It seems puzzling over how to assess different cell populations and molecules that dynamically interplay in the continuously altered microenvironment of inflamed intestinal mucosa of UC.
On the other hand, in the new “omics era”, high-throughput, discovery-driven approaches such as genomics, transcriptomics, proteomics and metabolomics have emerged providing a deeper and more comprehensive analysis of molecular biology of UC (5-8). Numerous bioinformatics-based tools have been developed to explore and integrate the huge amount of data generated by multi-“omics” profiling, aiming at characterizing the molecular “UC interactome” (Figure 1).
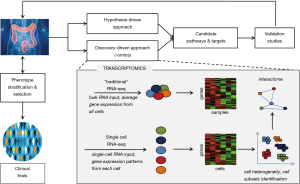
Recently, Smillie et al. (9), used a novel approach by applying single-cell RNA sequencing (scRNA-seq) analysis in colonic mucosa biopsies obtained from 18 active UC patients and 12 healthy individuals. Using sophisticated bioinformatics models and appropriate validation studies they provided a unique atlas of colonic mucosa in UC comprised of 366,650 single-cell transcriptomes that characterize the molecular signature of 51 different cell subsets.
Single-cell suspensions from fresh colonic biopsies were deployed after separation of the epithelial layer from the underlying lamina propria. Obtained cells were initially partitioned into epithelial, stromal and immune compartments for each sample, a process followed by clustering into distinct subsets at multi-sample level; all clusters were scored for certain gene signatures of interest. Their results illustrated for the first time that specific cell types and delineated pathways are linked with UC and drug susceptibility to the disease. In particular, microfold-like cells, inflammatory monocytes and fibroblasts, and CD8+IL-17+ T cells were found to highly expanded and characterized inflamed mucosa of active UC. Moreover, these subsets were in the center of the main cell-cell interaction networks, indicating that may mediate signals between diverse cell populations. In these networks, alterations in expression of genes that govern cell-cell interactions (i.e., receptors/counter receptors) may predict the dominant cellular compartments during different stages of intestinal inflammation. However, it has to be taken in consideration that neutrophil subsets were excluded from scRNA-seq analysis due to limitations of the tissue cells isolation protocol used. Several studies have demonstrated that neutrophils appear to play an early and central pathogenic role in active UC. Disease activity is correlated with neutrophil infiltration in inflamed mucosa and increased numbers of circulating neutrophils (10-12). Currently, growing evidence supports that neutrophils exhibit substantial plasticity that permits their phenotypic and functional adaption making them highly heterogeneous (13-15). Accordingly, neutrophil subsets with potentially important differences in function have been described and associated with disease phenotype in several inflammatory diseases including UC (16,17).
Comparison between non-inflamed and inflamed UC epithelial tissue uncovered also links between metabolic and inflammatory pathways. Interestingly, inflamed enterocytes were found to induce the hypoxia-inducible factor-1 (HIF-1) pathway, a well-known up-stream inducer of REDD1/autophagy axis that has recently described to be involved in tissue inflammation in active UC (17).
Another finding of interest that deserves a comment is the induction of a pro-inflammatory IL-17 response in CD8+ T cell subsets. How IL-17 is implicated in the inflammation of UC is not clarified yet; however clinical observations have indicated that anti-IL-17 treatment is associated with IBD exacerbation (18). Whether IL-17 response is protective or harmful, and whether this phenomenon is time-dependent across disease evolution remains to be explored.
Regarding resistance to anti-TNF treatment, authors examined the expression of oncostatin M (OSM), a target that had been previously linked with poor response to TNF inhibition (19), revealing inflammation associated fibroblasts (IAFs) as the most enriched OSM receptor gene cell subset. Given that IAFs were found to be among the colon tissue cells that highly expanded in patients refractory to anti-TNF therapies, authors suggested that IAFs may be implicated in drug resistance in UC by compensating TNF blockade. This finding provides novel research insights for the design of experimental studies concerning the unmet clinical need of refractory UC. Moreover, the authors proposed that many UC risk genes are cell type specific and co-regulated within limited gene modules, thus providing clear-cut suggestions for the limited sets of cell types and pathways that interplay in disease pathophysiology. The latter observations lead to nominate specific risk genes across previous released GWAS loci for certain functions and roles.
scRNA-seq represents a delicate and efficient technique that permits the dissection of function and structure at the lowest level possible concomitantly, enabling deeper investigation of inextricably intertwined pathological mechanisms. Integration and interpretation of results has been achieved through hierarchical clustering, an elegant method for analyzing gene expression data by building clusters of genes with similar patterns of expression. This process is achieved by iteratively grouping together genes that are highly correlated in their expression matrix.
Intra- and inter-subject variability was efficiently manipulated to attenuate both type I and type II errors through gene grouping into bins and formation of a consensus list of 1,500 genes of interest after correction for “batch effects”, namely the cell clustering by patient or disease phenotype rather than by cell type or state.
The study of Smillie et al. (9) aims to reconstitute the pathological milieu within which the cataract of disease driven inflammatory process begins and unravels. Elaborate and efficient techniques have permitted converge of more than half of candidate risk genes for UC onto a handful of groups (“meta-nodules” using the clustering terminology). These gene groups correspond to certain GWAS loci. However, this approach may fail to identify risk genes that: (I) are expressed at a lower level or at selected patients—a paradigm can be found at the increased frequency of MEFV mutations observed in UC patients, especially in those with episodic arthritis, thus suggesting a possible modifying effect of MEFV in the disease process; the difference in distribution of MEFV exon 2 haplotypes between UC patients and both FMF patients and normal individuals, suggests that UC patients constitute a genetically distinct population (20), (II) are expressed at cells or tissues that are not profiled, for example neutrophils, a main component of UC inflamed mucosa and indicator of disease severity (10-12), as mentioned above, and (III) are not co-expressed with other genes so as to be detected through clustering. Furthermore, the information driven represent a snapshot—an analogue of cross-sectional study at the clinical level—as it totally fails to incorporate time; it is to our strong belief that the disease process is a dynamic phenomenon where input and output are both time-dependent. This is further supported (or could be explained) by the remitting/relapsing character of the disease, as well as the fact that mucosal inflammation, even asymptomatic, may precedes systemic and/or extraintestinal manifestations (1,2). Finally, since investigators did not study therapy-naïve patients, the effect of corticosteroids and other immunomodulatory drugs including biological agents in the reported transcriptional changes is unknown and cannot be ignored. To overcome this obstacle, Dynamic Hierarchical Clustering could be a future target; this approach, successfully introduced in developmental gene expression data by enabling the identification of samples or genes which are influential at different stages of development, surpasses the restrictive assumption of a fixed hierarchy and enables working with data generated over a period of time (21).
Smillie et al. (9) provide a unique single-cell transcriptome map of the healthy and inflamed UC colon that is expected to constitute a road-map for future functional, experimental and clinical studies. Undoubtedly, represents an excellent paradigm of how novel, cutting-edge technologies can be successfully implemented in the analysis of complex diseases of unknown pathogenesis. Today we are incessantly supported by the benefits of novel artificial intelligence and machine learning tools for identifying new knowledge while sustaining clinical research and decision making. Integrating “omics” in the context of system biology approaches will hopefully pave the way for the identification of novel biotargets for personalized diagnosis and treatment of complex, multifactorial, inflammatory diseases such as UC. Apparently, this will also facilitate the better stratification and selection of various, and many times overlapped, clinical phenotypes, which remain the crucial initial steps for translation research studies in medicine. However, carefully designed clinical trials are needed to evaluate the accumulated information (Figure 1).
Acknowledgments
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Ungaro R, Mehandru S, Allen PB, et al. Ulcerative colitis. Lancet 2017;389:1756-70. [Crossref] [PubMed]
- Magro F, Gionchetti P, Eliakim R, et al. European Crohn’s and Colitis Organisation [ECCO]. Third European Evidence-based Consensus on Diagnosis and Management of Ulcerative Colitis. Part 1: Definitions, Diagnosis, Extra-intestinal Manifestations, Pregnancy, Cancer Surveillance, Surgery, and Ileo-anal Pouch Disorders. J Crohns Colitis 2017;11:649-70. [Crossref] [PubMed]
- de Souza HS, Fiocchi C. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol 2016;13:13-27. [Crossref] [PubMed]
- Park JH, Peyrin-Biroulet L, Eisenhut M, et al. IBD immunopathogenesis: A comprehensive review of inflammatory molecules. Autoimmun Rev 2017;16:416-26. [Crossref] [PubMed]
- Jin L, Li L, Hu C, et al. Integrative Analysis of Transcriptomic and Proteomic Profiling in Inflammatory Bowel Disease Colon Biopsies. Inflamm Bowel Dis 2019;25:1906-18. [Crossref] [PubMed]
- Zhou H, Xi L, Ziemek D, et al. Molecular Profiling of Ulcerative Colitis Subjects from the TURANDOT Trial Reveals Novel Pharmacodynamic/Efficacy Biomarkers. J Crohns Colitis 2019;13:702-13. [Crossref] [PubMed]
- Planell N, Masamunt MC, Leal RF, et al. Usefulness of Transcriptional Blood Biomarkers as a Non-invasive Surrogate Marker of Mucosal Healing and Endoscopic Response in Ulcerative Colitis. J Crohns Colitis 2017;11:1335-46. [Crossref] [PubMed]
- Rantalainen M, Bjerrum JT, Olsen J, et al. Integrative transcriptomic and metabonomic molecular profiling of colonic mucosal biopsies indicates a unique molecular phenotype for ulcerative colitis. J Proteome Res 2015;14:479-90. [Crossref] [PubMed]
- Smillie CS, Biton M, Ordovas-Montanes J, et al. Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 2019;178:714-730.e22. [Crossref] [PubMed]
- Brazil JC, Louis NA, Parkos CA. The role of polymorphonuclear leukocyte trafficking in the perpetuation of inflammation during inflammatory bowel disease. Inflamm Bowel Dis 2013;19:1556-65. [Crossref] [PubMed]
- Bressenot A, Salleron J, Bastien C, et al. Comparing histological activity indexes in UC. Gut 2015;64:1412-8. [Crossref] [PubMed]
- Park S, Abdi T, Gentry M, et al. Histological Disease Activity as a Predictor of Clinical Relapse Among Patients With Ulcerative Colitis: Systematic Review and Meta-Analysis. Am J Gastroenterol 2016;111:1692-701. [Crossref] [PubMed]
- Silvestre-Roig C, Hidalgo A, Soehnlein O. Neutrophil heterogeneity: implications for homeostasis and pathogenesis. Blood 2016;127:2173-81. [Crossref] [PubMed]
- Scapini P, Marini O, Tecchio C, et al. Human neutrophils in the saga of cellular heterogeneity: insights and open questions. Immunol Rev 2016;273:48-60. [Crossref] [PubMed]
- Jablonska J, Granot Z. Neutrophil, quo vadis? J Leukoc Biol. 2017;102:685-688. [Crossref] [PubMed]
- Zhou G, Yu L, Fang L, et al. CD177(+) neutrophils as functionally activated neutrophils negatively regulate IBD. Gut 2018;67:1052-63. [Crossref] [PubMed]
- Angelidou I, Chrysanthopoulou A, Mitsios A, et al. REDD1/Autophagy Pathway Is Associated with Neutrophil-Driven IL-1β Inflammatory Response in Active Ulcerative Colitis. J Immunol 2018;200:3950-61. [Crossref] [PubMed]
- Abraham C, Dulai PS, Vermeire S, et al. Lessons Learned From Trials Targeting Cytokine Pathways in Patients With Inflammatory Bowel Diseases. Gastroenterology 2017;152:374-388.e4. [Crossref] [PubMed]
- West NR, Hegazy AN, Owens BMJ, et al. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med 2017;23:579-89. [Crossref] [PubMed]
- Giaglis S, Mimidis K, Papadopoulos V, et al. Increased frequency of mutations in the gene responsible for familial Mediterranean fever (MEFV) in a cohort of patients with ulcerative colitis: evidence for a potential disease-modifying effect? Dig Dis Sci 2006;51:687-92. [Crossref] [PubMed]
- Fowler A, Menon V, Heard NA. Dynamic Bayesian clustering. J Bioinform Comput Biol 2013;11:1342001. [Crossref] [PubMed]
Cite this article as: Skendros P, Papadopoulos V, Ritis K. A gene expression map of colon tissue in ulcerative colitis: new methods rewrite old stories. Biotarget 2020;4:2.


