
Bronchioalveolar stem cells vindicated!
Introduction
Fourteen years ago, Kim et al. (1) reported the existence of bronchioalveolar stem cells (BASCs) in the lung. These cells expressed both secretoglobin family 1A member 1 (Scgb1a1) and surfactant protein c (Sftpc), the differentiation marker of club cells and alveolar epithelial type 2 cells, respectively. The recent lineage tracing of these cells from the Zhou lab using an elegant genetic approach to permanently express a fluorescent marker in these double positive cells, demonstrated that these cells are indeed capable of self-renewing and of differentiating into either bronchiolar or alveolar epithelial cells upon injury, in vivo (2). This work is a large step forward in the lung field, demonstrating the biological relevance of the BASCs in the context of regeneration. This work opens the way to elucidating the basic mechanisms controlling the proliferation and differentiation processes at work in BASCs during repair after injury. It will also facilitate a better appreciation of the importance of these cells compared to other epithelial stem cells in the distal lung, such as alveolar type 2 (AT2) cells, the variant of club cells, or the lineage negative epithelial progenitor cells (expressing α6β4 integrin).
BASCs are critical for both cancer and regeneration
In 2005, Kim et al. proposed that stem cells expressing both Scgb1a1 and Sftpc, located at the bronchioalveolar duct junction (BADJ) could give rise to adenocarcinomas (1). These stem cells can be enriched by FACS on the basis of Sftpc, Scgb1a1, Sca1, and CD34 expression (as well as negative for CD31 and CD45) and self-renewed as well as differentiated (into club and alveolar cells) in vitro. Expression of oncogenic protein K-ras in these cells drastically enhanced their proliferation (3). As BASCs display self-renewal capabilities, acquisition of deleterious mutations in these cells could lead to tumor formation. Therefore, it was proposed that these cells could give rise to cancer stem cells. Interestingly, these cells often retain the differentiation markers of the primary tumor. Now that the lineage tracing of these cells has been achieved, the impact of activating oncogenes in these cells should be easily investigated. It was also reported that both human and murine lung tumor cells, transferred via intratracheal injection into the lung, led to secondary tumors that grew selectively in the BADJ (4).
In the context of lung regeneration, two models were used in the Liu et al. study (2). First, a single naphthalene intraperitoneal injection was used to trigger the ablation of all club cells (expressing high levels of cytP450), except for the variant of club cells (which expresses low levels of cytP450), lining the bronchial surface and terminal bronchioles. These survival cells are located close to neuroendocrine cells, which are found mostly within the large proximal airways and are organized in clusters referred to as neuroendocrine bodies (5). Variant of club cells likely play a minor role in terminal bronchiole repair, due to the fact that the neuroendocrine bodies are quite rare in the distal lung. Interestingly, the existence of club cells which also expressed Sftpc in the BADJ was already reported in 2002 (6). These cells were resistant to naphthalene treatment and expanded after injury, suggesting that they could be progenitors for both alveolar and bronchiolar cells.
In the second model of lung injury, bleomycin intratracheal exposure targeted the alveolar epithelial cells. The alveolar epithelium is composed of alveolar type I (AT1) and II (AT2) cells. AT2 cells secrete surfactants critical for reducing alveolar surface tension, thereby preventing the collapse of alveoli, whereas the flattened AT1 cells are instrumental in facilitating gas exchange between the blood and the air inside the lung. Numerous stromal cells are also present in the lung, and these cells are likely to support the different epithelial stem cells described therein, including the BASCs.
Use of a dual-recombinase genetic model to target the BASCs
The Zhou lab made elegant use of the previously described inducible Dre-ERT2 and Cre-ERT2 recombinases to lineage-label the BASCs. For this purpose, they generated a new Sftpc-DreERT2 line, and a new knock-in reporter line. These tools enabled, after combined Cre (driven by the previously described Scgb1a1-CreERT2 mice) and Dre activity, to constitutively express, from the Rosa26 promoter, a fluorescent reporter.
Cre and FLP are two tyrosine site-specific recombinases (SSRs) commonly used to recombine genomic DNA in vivo, and have become essential tools for biological research. While there are many options available for Cre-mediated lineage analyses, fewer options exist which use FLP recombinase; whereas tools employing the other SSRs are relatively rare. A report from the Stewart lab demonstrated that a new recombinase, called Dre, specifically recognized rox sites (in the same way Cre recognizes loxP sites) (7). Another improvement occurred when the Dre was fused to the mutant estrogen receptor ERT2, allowing Dre regulation in a cell specific and temporal fashion (8).
The arsenal of SSRs is therefore expanding: first the Cre, then the FLP, and now the Dre as well as Vika. These recombinases recognize specific sites on the DNA (loxP, frt, rox, and vox, respectively) (Figure 1A). Interestingly, a polyvalent single reporter mouse line, which responds to the activity of each of these different recombinases, was recently reported, and will likely be instrumental as other mouse models are developed (9).
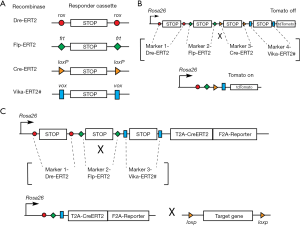
While it is a welcomed improvement to be able to label cells based on the expression of two markers, we potentially have a more powerful tool at our disposal to specifically label cells in vivo based on the expression of four markers. A new reporter line containing, for example, successive loxP-STOP-loxP, frt-STOP-frt, rox-STOP-rox, vox-STOP-vox cassettes proceeding a fluorescent marker could be generated (Figure 1B). For lineage tracing experiments, the fluorescent marker would be activated in only those very specific cells expressing the four recombinases. More importantly, powerful experimental models can be developed using multi-driver recombination to functionally activate or remove genes in targeted cells. As the majority of existing conditional alleles are mostly based on the Cre-loxP system, a superb approach would be to generate a Rosa26-based knock-in mouse with frt-STOP-frt, rox-STOP-rox, vox-STOP-vox cassettes preceding a CreERT2 and fluorescent reporter (Figure 1C). In this way, CreERT2, along with a reporter, would be expressed only when the three recombinases are present and have recombined their respective STOP cassettes. The CreERT2 could then be activated to induce DNA recombination (leading to gain or loss of function of a specific gene) in a given cell type. This technology, which is still in its infancy, is indeed very promising.
How similar are the BASCs to multipotent epithelial progenitor cells during early lung development?
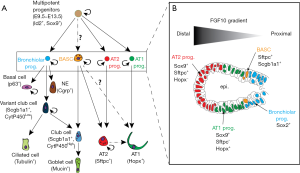
Multipotent lung epithelial cells expressing Id2 and Sox9 give rise to alveolar and bronchiolar lineages (Figure 2A). We have recently found that these cells are dependent upon fibroblast growth factor 10 (Fgf10) for the maintenance of their differentiation along the alveolar lineage (10) (Figure 2B). Upon loss of Fgf10 signaling, these cells acquire Sox2 expression, a marker of the bronchiolar lineage. Now that a lineage tracing strategy of the BASCs has been established, it will be important to determine when, during embryonic development, these cells are emerging. For example, are these cells emerging from the Sox9 and Sftpc-positive alveolar progenitors which acquire, later on, Scgb1a1? Or are they emerging from the Sox2 and Scgb1a1-positive cells and later acquire Sftpc? Interestingly, we have previously demonstrated that in the adult lung, the BASCs are amplified when Fgf10 is overexpressed in the lung (11), suggesting that these cells could respond to Fgf signals [in line with this idea, Liu et al. show that Fgfr2 expression was upregulated in BASCs, based on the single-cell RNA sequencing (2)]. It remains to be shown whether BASCs specifically express the Fgfr2b isoform, the main Fgf10 receptor, which would suggest a direct effect of Fgfs on BASCs. Lineage tracing of these cells combined with single cell RNA-seq in the context of either naphthalene or bleomycin injury will be able to identify the different relevant pathways activated in these cells during the repair process.
Acknowledgments
Funding: S Bellusci was supported by grants from the Deutsche Forschungsgemeinschaft (DFG; BE4443/1-1, BE4443/4-1, BE4443/6-1, BE4443/14-1, KFO309 P7 and SFB1213- projects A02 and A04), UKGM, Universities of Giessen and Marburg Lung Center (UGMLC) and the Deutsche Zentrum fur Lungenforschung (DZL).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by Executive Editor-in-Chief Dr. Hualin Sun (Jiangsu Key Laboratory of Neuroregeneration, Nantong University, Nantong, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/biotarget.2019.04.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kim CF, Jackson EL, Woolfenden AE, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 2005;121:823-35. [Crossref] [PubMed]
- Liu Q, Liu K, Cui G, et al. Lung regeneration by multipotent stem cells residing at the bronchioalveolar-duct junction. Nat Genet 2019;51:728-38. [Crossref] [PubMed]
- Jackson EL, Willis N, Mercer K, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 2001;15:3243-8. [Crossref] [PubMed]
- Kim CF. Paving the road for lung stem cell biology: bronchioalveolar stem cells and other putative distal lung stem cells. Am J Physiol Lung Cell Mol Physiol 2007;293:L1092-8. [Crossref] [PubMed]
- Reynolds SD, Hong KU, Giangreco A, et al. Conditional clara cell ablation reveals a self-renewing progenitor function of pulmonary neuroendocrine cells. Am J Physiol Lung Cell Mol Physiol 2000;278:L1256-63. [Crossref] [PubMed]
- Giangreco A, Reynolds SD, Stripp BR. Terminal bronchioles harbor a unique airway stem cell population that localizes to the bronchoalveolar duct junction. Am J Pathol 2002;161:173-82. [Crossref] [PubMed]
- Anastassiadis K, Fu J, Patsch C, et al. Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice. Dis Model Mech 2009;2:508-15. [Crossref] [PubMed]
- Devine WP, Wythe JD, George M, et al. Early patterning and specification of cardiac progenitors in gastrulating mesoderm. Elife 2014; [Crossref] [PubMed]
- Karimova M, Baker O, Camgoz A, et al. A single reporter mouse line for Vika, Flp, Dre, and Cre-recombination. Sci Rep 2018;8:14453. [Crossref] [PubMed]
- Jones MR, Dilai S, Lingampally A, et al. A Comprehensive Analysis of Fibroblast Growth Factor Receptor 2b Signaling on Epithelial Tip Progenitor Cells During Early Mouse Lung Branching Morphogenesis. Front Genet 2019;9:746. [Crossref] [PubMed]
- Volckaert T, Dill E, Campbell A, et al. Parabronchial smooth muscle constitutes an airway epithelial stem cell niche in the mouse lung after injury. J Clin Invest 2011;121:4409-19. [Crossref] [PubMed]
Cite this article as: Jones M, Zhang JS, Bellusci S. Bronchioalveolar stem cells vindicated! Biotarget 2019;3:4.


